AORC Data#
DayMet’s API has recently gone down, leading us to experiment with a new type of met data, AORC, which is the dataset used to drive the National Water Model.
%load_ext autoreload
%autoreload 2
AORC provides gridded meteorological data for the conterminous US and Alaska at ~800m spatial resolution with hourly timestep from 1979 (Conterminous US) or 1981 (Alaska) ~ present. website
Available variables:
Variable |
Description (units) |
|---|---|
APCP_surface |
Total precipitation [mm] |
DLWRF_surface |
Incident longwave radiation flux density [W m^-2] |
DSWRF_surface |
Incident shortwave radiation flux density [W m^-2] |
PRES_surface |
Air pressure [Pa] |
SPFH_2maboveground |
Specific Humidity [g/g] |
TMP_2maboveground |
Air temperature [°C] |
UGRD_10maboveground |
West-East component of the wind velocity [m s^-1] |
VGRD_10maboveground |
North-South component of the wind velocity [m s^-1] |
Notes:
AORC, unlike DayMet, provides all Gregorian calendar days
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['figure.dpi'] = 150
import logging
import numpy as np
import os
import cftime, datetime
import watershed_workflow
import watershed_workflow.ui
import watershed_workflow.sources
import watershed_workflow.meteorology
import watershed_workflow.io
watershed_workflow.ui.setup_logging(1,None)
coweeta_shapefile = 'Coweeta/input_data/coweeta_basin.shp'
crs = watershed_workflow.crs.default_crs
Load the Watershed#
coweeta_source = watershed_workflow.sources.ManagerShapefile(coweeta_shapefile, id_name='BASIN_CODE')
coweeta = coweeta_source.getShapes(out_crs=crs)
watershed = watershed_workflow.SplitHUCs(coweeta)
watershed_workflow.split_hucs.simplify(watershed, 60)
Download the raw DayMet raster#
returned raw data has dim(nband, ncol, nrow)
startdate = "2010-1-1"
enddate = "2013-12-31"
# setting vars = None to download all available variables
source = watershed_workflow.sources.ManagerAORC()
met_data = source.getDataset(coweeta, start=startdate, end=enddate)
Variable size: 79.2 TB
<xarray.Dataset> Size: 79TB
Dimensions: (time: 35064, latitude: 4201, longitude: 8401)
Coordinates:
* latitude (latitude) float64 34kB 20.0 20.01 20.02 ... 54.99 55.0
* longitude (longitude) float64 67kB -130.0 -130.0 ... -60.01 -60.0
* time (time) datetime64[ns] 281kB 2010-01-01 ... 2013-12-3...
Data variables:
APCP_surface (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
DLWRF_surface (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
DSWRF_surface (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
PRES_surface (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
SPFH_2maboveground (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
TMP_2maboveground (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
UGRD_10maboveground (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
VGRD_10maboveground (time, latitude, longitude) float64 10TB dask.array<chunksize=(144, 128, 256), meta=np.ndarray>
Subsetting:
lon: (np.float64(-83.4868), np.float64(-83.4133))
lat: (np.float64(35.019), np.float64(35.0822))
Variable size: 0.142 GB
Warp the dataset#
AORC comes back as a non-projected dataset in lat-lon. We need to warp it to a projected dataset for use in ATS.
met_data_warped = watershed_workflow.warp.dataset(met_data, crs, 'bilinear')
Save to Disk#
NetCDF file#
By default, the raw data is saved to $DATA_DIR/meteorology/aorc/ in a netcdf file.
Write to ATS format#
This will write daymet in a format that ATS can read. E.g., this will partition precipitation into rain and snow, convert vapor pressure to relative humidity, get mean air temperature and so on.
dout has dims of
(ntime, nrow, ncol)or(ntime, ny, nx)
# convert the data to ATS units, partitioning precip into rain and snow, and computing daily averages
met_data_ats = watershed_workflow.meteorology.convertAORCToATS(met_data_warped)
filename = os.path.join('Coweeta', 'output_data', 'watershed_daymet-ats.h5')
watershed_workflow.io.writeDatasetToHDF5(
filename,
met_data_ats,
met_data_ats.attrs)
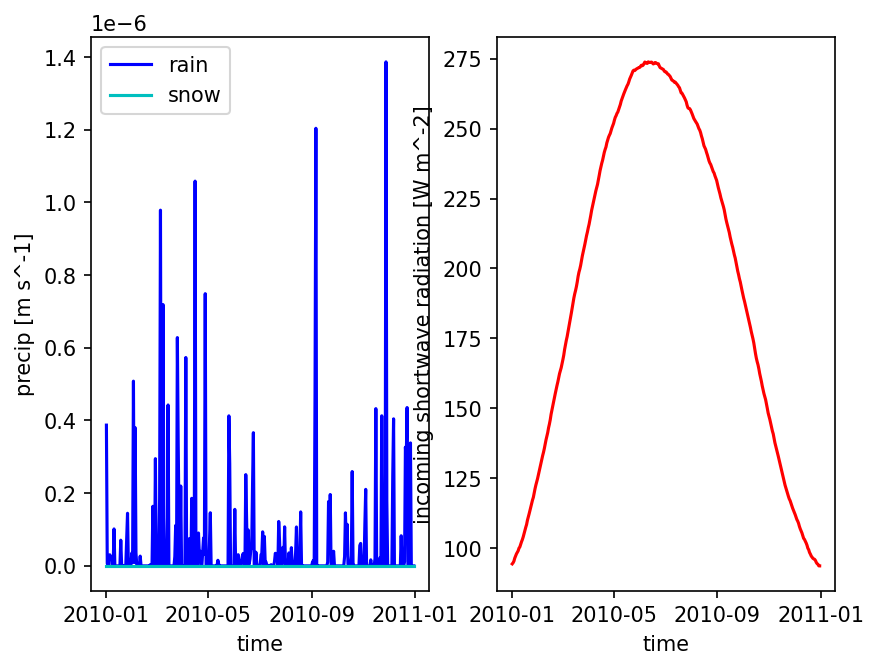
# plot one pixel as a function of time
fig = plt.figure()
ax = fig.add_subplot(121)
met_data_single_pixel = met_data_ats.isel({'time':slice(0,365),
'x' : 5,
'y' : 5})
met_data_single_pixel['precipitation rain [m s^-1]'].plot(color='b', label='rain')
met_data_single_pixel['precipitation snow [m SWE s^-1]'].plot(color='c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.set_title('')
ax.legend()
ax = fig.add_subplot(122)
met_data_single_pixel['incoming shortwave radiation [W m^-2]'].plot(color='r', label='qSW_in')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
ax.set_title('')
plt.show()
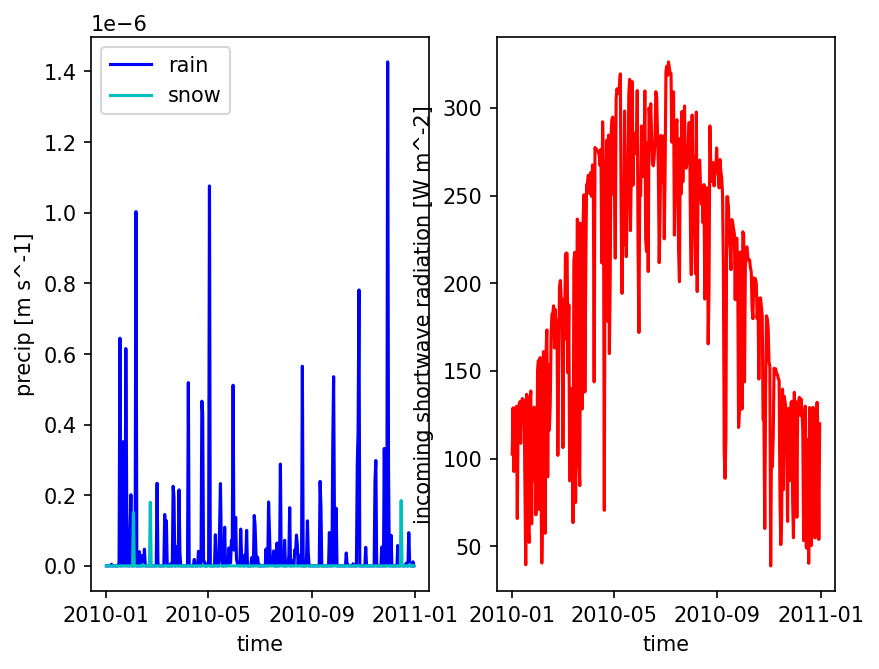
Smooth to form a typical year#
A “typical” year is commonly used in a cyclic, annual steady-state spinup. We form this by averaging all Jan 1s in the record, averaging all Jan 2s, etc.
met_data
<xarray.Dataset> Size: 142MB
Dimensions: (time: 35041, latitude: 7, longitude: 9)
Coordinates:
* latitude (latitude) float64 56B 35.02 35.03 ... 35.07 35.07
* longitude (longitude) float64 72B -83.49 -83.48 ... -83.43 -83.42
* time (time) object 280kB 2010-01-01 00:00:00 ... 2013-12-...
Data variables:
APCP_surface (time, latitude, longitude) float64 18MB ...
DLWRF_surface (time, latitude, longitude) float64 18MB ...
DSWRF_surface (time, latitude, longitude) float64 18MB ...
PRES_surface (time, latitude, longitude) float64 18MB ...
SPFH_2maboveground (time, latitude, longitude) float64 18MB ...
TMP_2maboveground (time, latitude, longitude) float64 18MB ...
UGRD_10maboveground (time, latitude, longitude) float64 18MB ...
VGRD_10maboveground (time, latitude, longitude) float64 18MB ...
Attributes:
name: AORC v1.1
source: NOAA AWS S3 Zarr# aggregate to daily data
met_data_daily = met_data.resample(time=datetime.timedelta(hours=24)).mean()
ndays = met_data_daily['APCP_surface'].shape[0]
print('nyears = ', ndays // 365)
print('n days leftover = ', ndays % 365)
print('note, that leftover day is a leap day')
nyears = 4
n days leftover = 1
note, that leftover day is a leap day
# cannot compute a typical year for leap year -- need to align days-of-the-year
# remove leap day (Dec 31 of leap years)
met_data_noleap = watershed_workflow.data.filterLeapDay(met_data_daily)
# smooth the raw data
met_data_smooth = watershed_workflow.data.smoothTimeSeries(met_data_noleap, method='savgol', window_length=181, polyorder=2)
# then compute a "typical" year
met_data_typical = watershed_workflow.data.computeAverageYear(met_data_smooth, 'time', 2010, 2)
# then reproject
met_data_typical_warped = watershed_workflow.warp.dataset(met_data_typical, crs, 'bilinear')
# and lastly convert to ATS
met_data_typical_ats = watershed_workflow.meteorology.convertAORCToATS(met_data_typical_warped)
# plot one pixel as a function of time
fig = plt.figure()
ax = fig.add_subplot(121)
met_data_single_pixel = met_data_typical_ats.isel({'time':slice(0,365),
'x' : 5,
'y' : 5})
met_data_single_pixel['precipitation rain [m s^-1]'].plot(color='b', label='rain')
met_data_single_pixel['precipitation snow [m SWE s^-1]'].plot(color='c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.set_title('')
ax.legend()
ax = fig.add_subplot(122)
met_data_single_pixel['incoming shortwave radiation [W m^-2]'].plot(color='r', label='qSW_in')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
ax.set_title('')
plt.show()
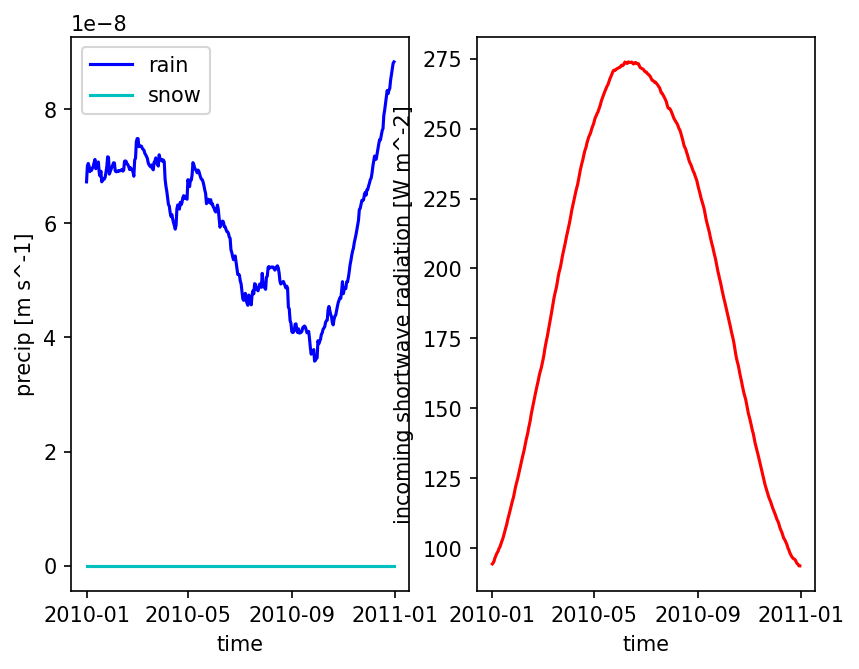
Other choices for Precip#
Often smoothing precip like that is a bad idea – you now have every day misting with low intensity rain which can result in a ton of interception and canopy evaporation and no transpiration. Another approach is to just take the median total rainfall year and repeat that year multiple times.
met_data_noleap['APCP_surface'].shape[0] % 365
0
precip_raw = met_data_noleap['APCP_surface']
shape_xy = precip_raw.shape[1:]
precip_raw = precip_raw.values.reshape((-1, 365,)+shape_xy)
annual_precip_raw = precip_raw.sum(axis=(1,2,3))
print(annual_precip_raw)
[4867.39590586 5486.27508175 5143.10007664 7351.33344288]
# find the median...
# note -- don't use np.median here... for even number of years it will not appear. Instead, sort and take the halfway point
median_i = sorted(((i,v) for (i,v) in enumerate(annual_precip_raw)), key=lambda x : x[1])[len(annual_precip_raw)//2][0]
spatially_averaged_typical_year_precip = precip_raw[median_i]
spatially_averaged_typical_year_precip.shape
(365, 7, 9)
# stuff the median year into the typical data, tiled for number of years being generated (here 2)
typical_precip_raw = np.tile(precip_raw[median_i], (2,1,1))
met_data_typical['APCP_surface'] = (['time', 'latitude', 'longitude'], typical_precip_raw)
# then reproject
met_data_typical_warped = watershed_workflow.warp.dataset(met_data_typical, crs, 'bilinear')
# convert that to ATS
met_data_typical_ats = watershed_workflow.meteorology.convertAORCToATS(met_data_typical_warped)
# plot one pixel as a function of time
fig = plt.figure()
ax = fig.add_subplot(121)
met_data_single_pixel = met_data_typical_ats.isel({'time':slice(0,365),
'x' : 5,
'y' : 5})
met_data_single_pixel['precipitation rain [m s^-1]'].plot(color='b', label='rain')
met_data_single_pixel['precipitation snow [m SWE s^-1]'].plot(color='c', label='snow')
ax.set_ylabel('precip [m s^-1]')
ax.set_title('')
ax.legend()
ax = fig.add_subplot(122)
met_data_single_pixel['incoming shortwave radiation [W m^-2]'].plot(color='r', label='qSW_in')
ax.set_ylabel('incoming shortwave radiation [W m^-2]')
ax.set_title('')
plt.show()